Chemical structures in chemistry files
Important chemistry files including chemical structures
| File extension | Origin | Conversion into .mol file | Example | converted into .mol file (not active yet…) |
|---|---|---|---|---|
| .cdx .cdxml | ChemDraw | OpenBabel | benzoic.cdx structure.cdxml | benzoic acid in .mol format |
| .log .out .g09 | Gaussia98/03/09/16 | OpenBabel | melezitose_betternmrJ.log | |
| .sdf | multiple open sources | OpenBabel | compound1.nmredata.sdf | |
| .mol | multiple open sources | no conversion | 2D .mol/.sdf (most frequent) | |
| .mol | multiple open sources | no conversion | 3D .mol/.sdf | |
| .mnova | MestreNova | Mnova | vaniline.mnova |
Visualisation can be done after conversion into the open .mol files format. The .mol file can be displayed using a specialized tools (nice for ) or as a fixed image after conversion of .mol into .png (or other format).
Distinguish 2D from 3D .mol (or .sdf) files.
OpenBabel
Conversion into .mol file using OpenBabel
obabel inputfile.sdf -O outputfile.mol
obabel inputfile.cdx -O outputfile.mol
obabel -ig03 inputfile.log -O outputfile.mol
Conversion into images using OpenBabel
obabel outputfile.mol -O outputfile.png
A separete web service returning the .mol or the .png when sending any other format would simplify interfacing and modularity.
Mnova
The Mnova software requires a licence. Extraction of elements from the .mnova files can be done using scripts called from unix:
/usr/bin/mnova "/usr/username/myScript.qs" -sf "myFunction",0.1,10,true,off // generic call of function from a for user-defined script
/usr/bin/mnova "NMReDATAExporter.qs" //for NMReDATA - this script my be a starting point for automated extraction. We may have to remove user interaction from this script.
babel -isdf 'mymols.cdx' -omol 'outputfile.mol' //if the data are not in .mol format already
A web service returning the .mol when sending a .mnova would simplify interfacing and modularity. When running scripts, the graphical interface is disabled.
Visualization of .mol files
| Extension | Source | Visualization tool | Demo |
|---|---|---|---|
| .mol | 2D structure | JSME | 2D structure of menthol (Preferred!) |
| .mol | 2D structure | Kekule | 2D structure of menthol |
| .mol | 3D structure | JSmol | 3D structure of cholesterol (Preferred!) |
Note about chemical structures in chemistry
In chemistry representation of the structure of componds is essential. We shall distinguish often called “2d” or “flat” structure that are drawing where 3D features are represented using special notation from full-flesh 3D structures where the X, Y, Z coordinates are specified.
2D structure
In most case, compounds are represented as a projection on the surface where they are drawn. The knowledge of the chemist makes it clear that the three bonds of the highligheted cabon are not on the same plane because a fourth bond with a hydrogen atom is implict. This is the reprentation of alanine. If the chemists wants to specify, which of the enantiomer he wants to represent, he uses a filled triangle or dashed triangle to indicate that the bonds is pointing up or down respectively. These represent L-alanine and D-alanine respectively.
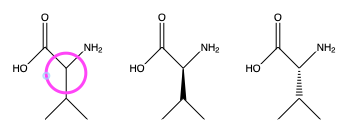
Converting a 2D structure into a 3D structure is not problematic when the stereochemistry is fully determined. If the type of alanine (D- or L-) is not fully determined (something that may reflect a genuine lack of information) it is better not to use 3D structures, or to generate all the possible structures and give the chemist the choice among them. It means that one should not systematically convert “2D” into “3d” structures - only do it when it is safe.
| alanine | InChI | InChIKey |
|---|---|---|
| DL-alanine | InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6) | QNAYBMKLOCPYGJ-UHFFFAOYSA-N |
| L-alanine | InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6)/t2-/m0/s1 | QNAYBMKLOCPYGJ-REOHCLBHSA-N |
| D-alanine | InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6)/t2-/m1/s1 | QNAYBMKLOCPYGJ-UWTATZPHSA-N |